MPDL Demonstrator Marine Microbiology
This is a protected page.
Participants[edit]
- MPDL
- Max-Planck-Institut für marine Mikrobiologie in Bremen see MPIMB
Short description[edit]
Use cases[edit]
- I would like to have a list of my publications linked to the Silva resources I have been working on represented on the home page of my research group and on my personal home page
- or in other wording: as a user of PubMan i would like to link my publication to one or more Silva resources via Silva accession ID, and have the Silva accession ID included into the PubMan citation
- I would like to be able to provide search by Silva accession ID and retrieve all publications related to this ID
- I would like to provide coordinates of a search area, and be able to retrieve a list of all publications and Silva resources related to Marine Microbiology expeditions in that area.
- I would like to have a list of all publications related to an expedition
- In my blog http://wissenschafts-blog.abendblatt.de I would like to provide an RSS feed of the publications that reference the expedition the blog is about
Demonstrator[edit]
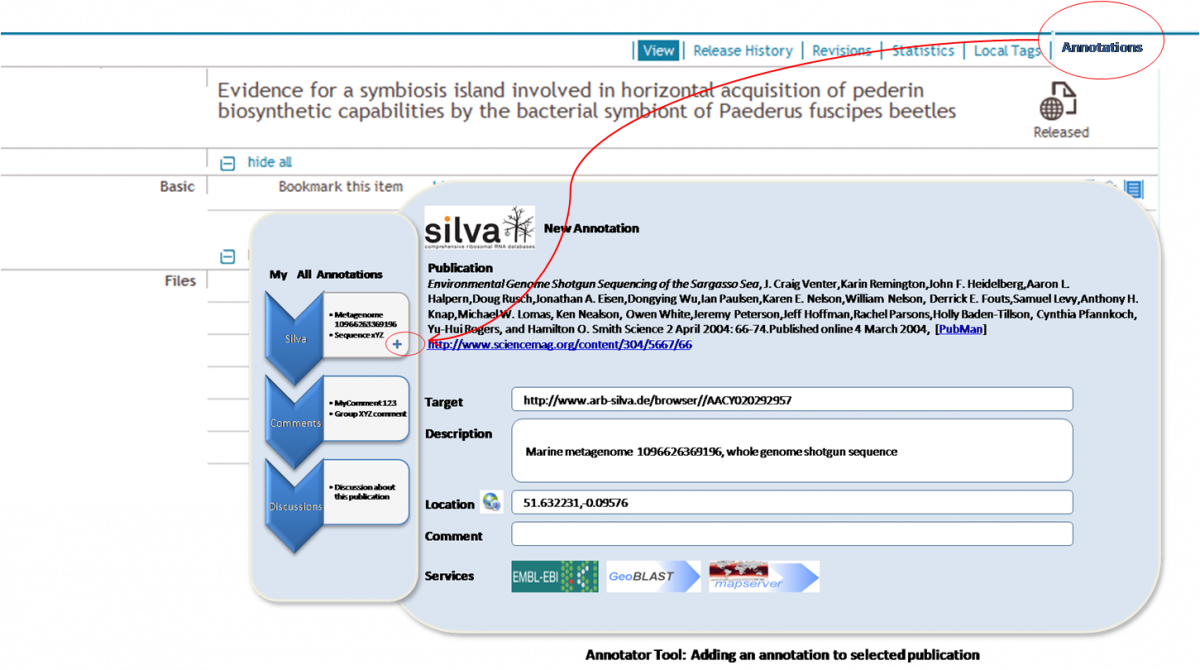
In the basic demonstration scenario, PubMan will be extended with a functionality to annotate a publication by linking it to external data resources of interest e.g. probes, genomes, meta-genomes etc. In the first stage the Silva database will be considered as a repository where such data resources reside. A screen Mock-up for the envisioned scenario is displayed in the image below (note that mock-up and implementation may differ after the revision with the Institute): .
Create new Silva Annotation[edit]
- User chooses to create a Silva Annotation for a particular Publication
- The system offers a form for entering an annotation (the form also contains a list of icons for available services for a Silva annotation e.g. GeoBlast, Map Server, EMBL-EBI)
- User enters a URL of the Silva resource
- The System fetches the Silva resource data from predefined web service (e.g. Silva service) and populates the annotation fields (e.g. description, location, etc.) correspondingly
- User adds additional metadata if necessary (e.g. comment) and saves the Annotation
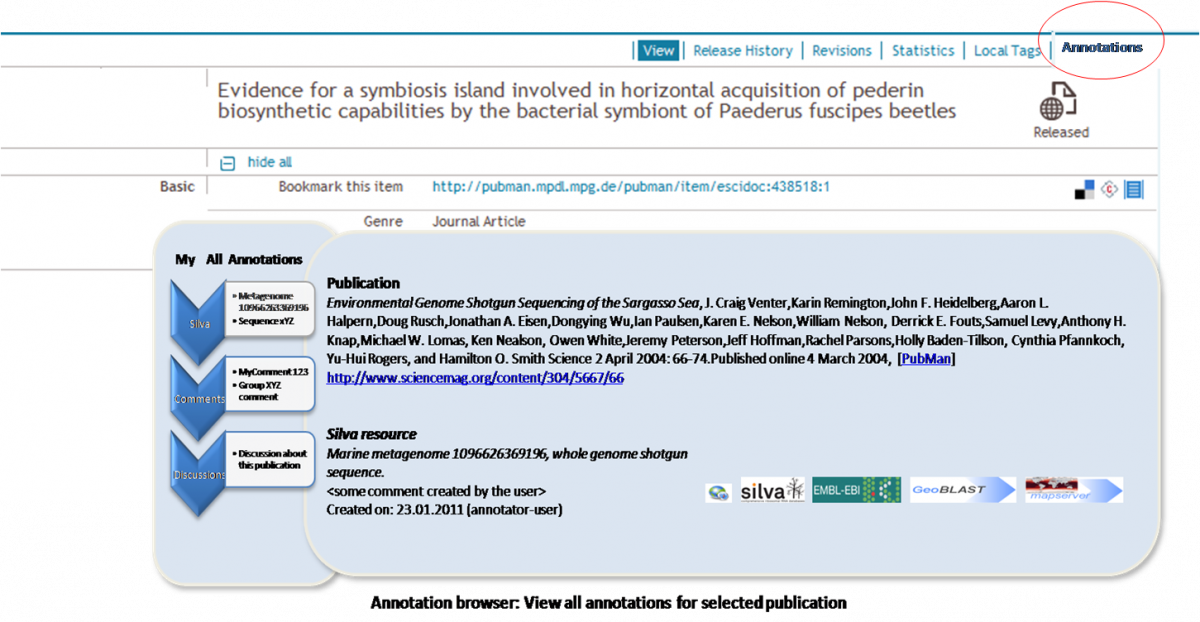
View Annotation[edit]
- User chooses to view Annotations for the Publication
- The System displays the Annotation Browser and lists all annotations for the selected publication
- The user may also browse through other types of Annotations that exist for the selected publication
- The user may decide to modify an annotation or add another annotation of particular annotation type
- The user may decide to invoke an external service for the target source (i.e. Silva resource) referenced in the annotation, by triggering respective service icon (e.g. GeoBlast, EMBL)
List my publications linked to the annotated resources for inclusion in my web pages[edit]
- User chooses to export his publications into a new "APA-annotated" citation style
- User defines the search criteria to get a list of publications
- The system provides an APA-annotated citation style (with additional information on annotated resources), as in the example below:
[edit]
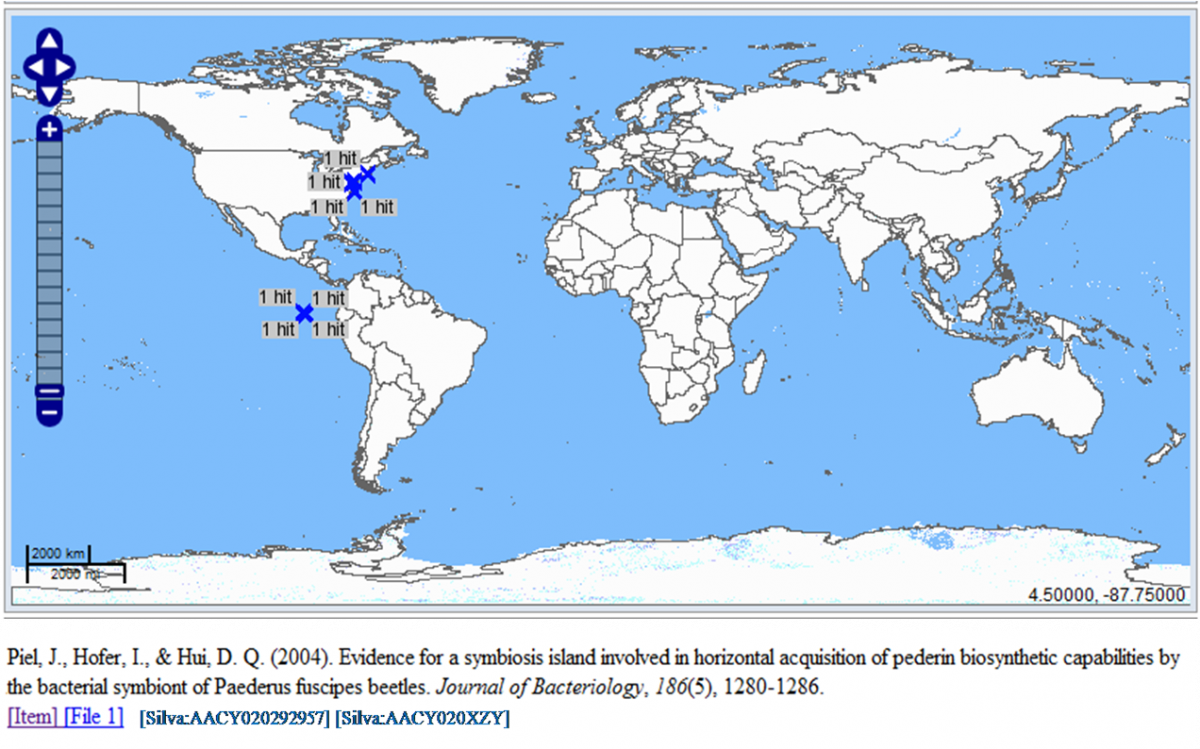
- User enters coordinates of a search area into the search interface(see Note below)
- The system retrieves a list of Silva sequences for the requested coordinates
- The system retrieves a list of publications for the requested Silva sequences
- The system displays a Map showing the hits for sequences and a list of related publications into a new "APA-annotated" citation style
Note: this use case may be used as an integration with the GeoBlast web service
Related services and information[edit]
Important things[edit]
- chemistry - analysis, analytical chemistry, nucleic acid analyses, genome analysis and metagenomics, field work, mathematical simulations, experimental visualizations
- Publications
- Expeditions
- Projects (experimental - field research and probes)
- Locations
- Facilities and methods
- RNA sequences for all three domains of life (Bacteria, Archaea and Eukarya)
- FISH and probes - online resource for information on the identification of individual microbial cells by fluorescence in situ hybridization (FISH) with ribosomal RNA-targeted oligonucleotide probes
- Protocols eg. http://www.arb-silva.de/fileadmin/graphics_fish/SILVA_FISH_protocols_fixation_100618.pdf
- Marine ecological genomics, integrating genomic, metagenomic and ribosomal RNA data with primary environmental data and curated metadata
- SILVA - on-line resource for quality checked and aligned ribosomal RNA sequence data, free for academic use
- contextual data, see also http://www.arb-silva.de/projects/contextual-data/
- contextual data, see also http://gensc.org/gc_wiki/index.php/MIENS
- Blogs from the expeditions and probes see http://wissenschafts-blog.abendblatt.de